
Simulated survival dataset with delayed treatment effect
Source:R/mb_delayed_effect.R
mb_delayed_effect.RdMagirr and Burman (2019) considered several scenarios for their
modestly weighted logrank test.
One of these had a delayed treatment effect with a hazard ratio
of 1 for 6 months followed by a hazard ratio of 1/2 thereafter.
The scenario enrolled 200 patients uniformly over 12 months and
cut data for analysis 36 months after enrollment was opened.
This dataset was generated by the sim_pw_surv() function
under the above scenario.
References
Magirr, Dominic, and Carl‐Fredrik Burman. 2019. "Modestly weighted logrank tests." Statistics in Medicine 38 (20): 3782–3790.
Examples
library(survival)
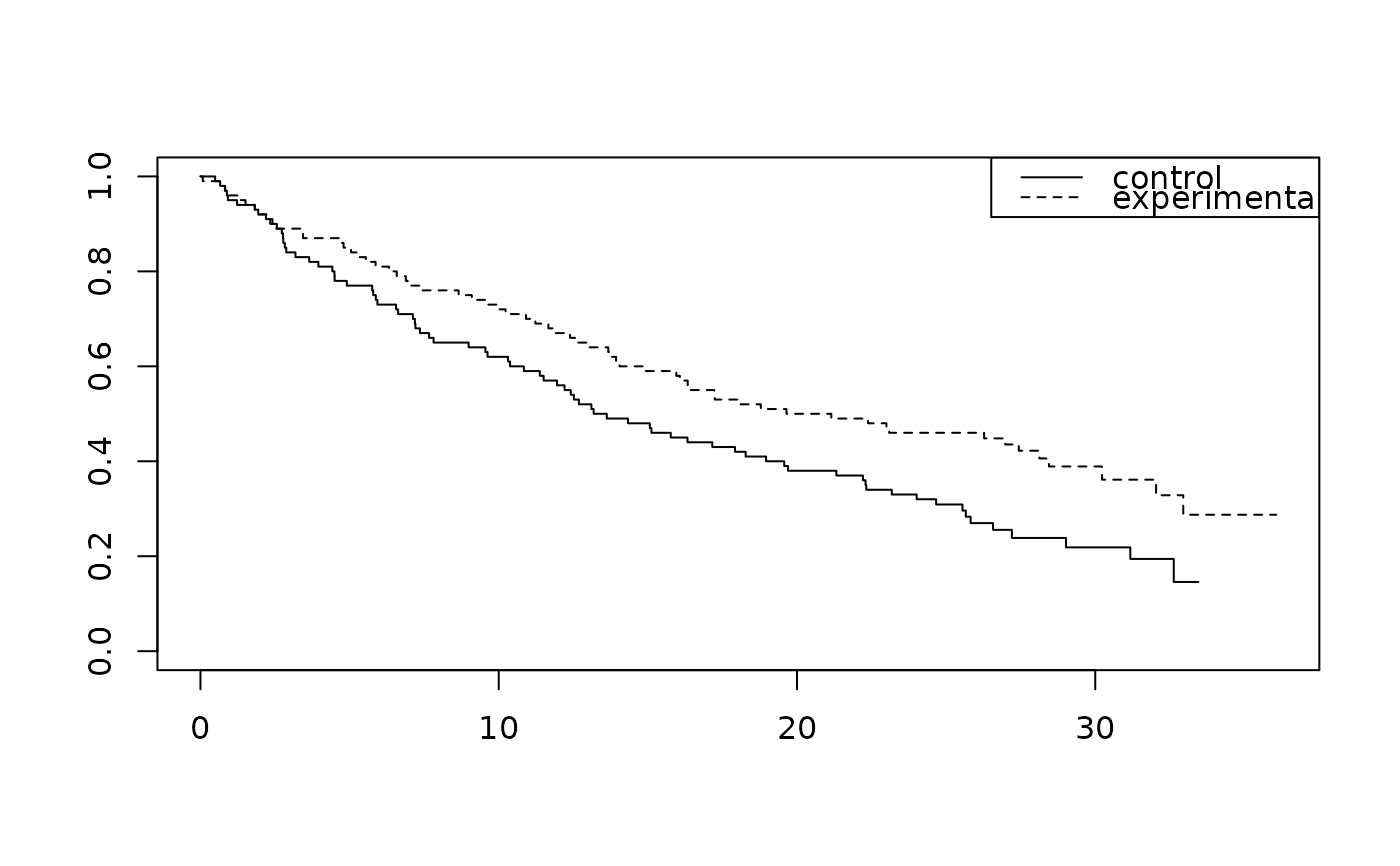
fit <- survfit(Surv(tte, event) ~ treatment, data = mb_delayed_effect)
# Plot survival
plot(fit, lty = 1:2)
legend("topright", legend = c("control", "experimental"), lty = 1:2)
 # Set up time, event, number of event dataset for testing
# with arbitrary weights
ten <- mb_delayed_effect |> counting_process(arm = "experimental")
head(ten)
#> stratum event_total event_trt tte n_risk_total n_risk_trt s
#> 1 All 1 1 0.07659251 200 100 1.000
#> 2 All 1 0 0.49067015 199 99 0.995
#> 3 All 1 1 0.65465035 198 99 0.990
#> 4 All 1 0 0.65906384 197 98 0.985
#> 5 All 1 1 0.81945349 196 98 0.980
#> 6 All 1 0 0.82788909 195 97 0.975
#> o_minus_e var_o_minus_e
#> 1 0.5000000 0.2500000
#> 2 -0.4974874 0.2499937
#> 3 0.5000000 0.2500000
#> 4 -0.4974619 0.2499936
#> 5 0.5000000 0.2500000
#> 6 -0.4974359 0.2499934
# MaxCombo with logrank, FH(0,1), FH(1,1)
mb_delayed_effect |>
maxcombo(rho = c(0, 0, 1), gamma = c(0, 1, 1), return_corr = TRUE)
#> $method
#> [1] "MaxCombo"
#>
#> $parameter
#> [1] "FH(0, 0) + FH(0, 1) + FH(1, 1)"
#>
#> $z
#> [1] -2.473248 -2.424018 -2.482653
#>
#> $corr
#> v1 v2 v3
#> 1 1.0000000 0.8606625 0.9312916
#> 2 0.8606625 1.0000000 0.9579831
#> 3 0.9312916 0.9579831 1.0000000
#>
#> $p_value
#> [1] 0.01104817
#>
# Generate another dataset
ds <- sim_pw_surv(
n = 200,
enroll_rate = data.frame(rate = 200 / 12, duration = 12),
fail_rate = data.frame(
stratum = c("All", "All", "All"),
period = c(1, 1, 2),
treatment = c("control", "experimental", "experimental"),
duration = c(42, 6, 36),
rate = c(log(2) / 15, log(2) / 15, log(2) / 15 * 0.6)
),
dropout_rate = data.frame(
stratum = c("All", "All"),
period = c(1, 1),
treatment = c("control", "experimental"),
duration = c(42, 42),
rate = c(0, 0)
)
)
# Cut data at 24 months after final enrollment
mb_delayed_effect_2 <- ds |> cut_data_by_date(max(ds$enroll_time) + 24)
# Set up time, event, number of event dataset for testing
# with arbitrary weights
ten <- mb_delayed_effect |> counting_process(arm = "experimental")
head(ten)
#> stratum event_total event_trt tte n_risk_total n_risk_trt s
#> 1 All 1 1 0.07659251 200 100 1.000
#> 2 All 1 0 0.49067015 199 99 0.995
#> 3 All 1 1 0.65465035 198 99 0.990
#> 4 All 1 0 0.65906384 197 98 0.985
#> 5 All 1 1 0.81945349 196 98 0.980
#> 6 All 1 0 0.82788909 195 97 0.975
#> o_minus_e var_o_minus_e
#> 1 0.5000000 0.2500000
#> 2 -0.4974874 0.2499937
#> 3 0.5000000 0.2500000
#> 4 -0.4974619 0.2499936
#> 5 0.5000000 0.2500000
#> 6 -0.4974359 0.2499934
# MaxCombo with logrank, FH(0,1), FH(1,1)
mb_delayed_effect |>
maxcombo(rho = c(0, 0, 1), gamma = c(0, 1, 1), return_corr = TRUE)
#> $method
#> [1] "MaxCombo"
#>
#> $parameter
#> [1] "FH(0, 0) + FH(0, 1) + FH(1, 1)"
#>
#> $z
#> [1] -2.473248 -2.424018 -2.482653
#>
#> $corr
#> v1 v2 v3
#> 1 1.0000000 0.8606625 0.9312916
#> 2 0.8606625 1.0000000 0.9579831
#> 3 0.9312916 0.9579831 1.0000000
#>
#> $p_value
#> [1] 0.01104817
#>
# Generate another dataset
ds <- sim_pw_surv(
n = 200,
enroll_rate = data.frame(rate = 200 / 12, duration = 12),
fail_rate = data.frame(
stratum = c("All", "All", "All"),
period = c(1, 1, 2),
treatment = c("control", "experimental", "experimental"),
duration = c(42, 6, 36),
rate = c(log(2) / 15, log(2) / 15, log(2) / 15 * 0.6)
),
dropout_rate = data.frame(
stratum = c("All", "All"),
period = c(1, 1),
treatment = c("control", "experimental"),
duration = c(42, 42),
rate = c(0, 0)
)
)
# Cut data at 24 months after final enrollment
mb_delayed_effect_2 <- ds |> cut_data_by_date(max(ds$enroll_time) + 24)