
Approximating an arbitrary survival distribution
Keaven M. Anderson
Source:vignettes/articles/story-arbitrary-distribution.Rmd
story-arbitrary-distribution.RmdIntroduction
We demonstrate how to approximate arbitrary continuous survival distributions with piecewise exponential approximations. This enables sample size computations for arbitrary survival models using software designed for the piecewise exponential distribution. Three functions in particular are demonstrated:
Lognormal approximation
We demonstrate s2pwe() by approximating the lognormal
distribution with piecewise exponential failure rates. Note that when
the resulting log_normal_rate is used, the final piecewise
exponential duration is extended. That is, we have
arbitrarily approximated with 6 piecewise exponential rates for a
duration of 1 unit of time (say, month) followed by a final rate which
extends to infinity.
log_normal_rate <- s2pwe(
times = c(1:6, 9),
survival = plnorm(c(1:6, 9), meanlog = 0, sdlog = 2, lower.tail = FALSE)
)
log_normal_rate## # A tibble: 7 × 2
## duration rate
## <dbl> <dbl>
## 1 1 0.693
## 2 1 0.316
## 3 1 0.224
## 4 1 0.177
## 5 1 0.148
## 6 1 0.128
## 7 3 0.103We compare the resulting approximation to the actual lognormal
survival using ppwe() to compute survival probabilities
\(P\{T>t\}\). For a better
approximation, use a larger number of points. We plot with a log scale
on the y-axis since the piecewise exponential survival from
ppwe() is piecewise linear on this scale. We note that at
the beginning of each rate period in the approximation the actual
survival distribution and its approximation match exactly as indicated
by the circles on the graph.
# Use a large number of points to plot lognormal survival
times <- seq(0, 12, .025)
plot(times, plnorm(times, meanlog = 0, sdlog = 2, lower.tail = FALSE),
log = "y", type = "l",
main = "Lognormal Distribution vs. Piecewise Approximation", yaxt = "n",
ylab = "log(Survival)", col = 1
)
# Now plot the pieceise approximation using the 7-point approximation from above
lines(
times,
ppwe(x = times, duration = log_normal_rate$duration, rate = log_normal_rate$rate),
col = 2
)
# Finally, add point markers at the points used in the approximation
points(x = c(0:6), plnorm(c(0:6), meanlog = 0, sdlog = 2, lower.tail = FALSE), col = 1)
text(x = c(5, 5), y = c(.5, .4), labels = c("Log-normal", "Piecewise Approximation (7 pts)"), col = 1:2, pos = 4)
We considered the lognormal distribution due to the flexibility it allows for hazard rates over time; see, for example, Wikipedia.
Poisson mixture model
We consider a Poisson mixture model to incorporate a cure model into sample size planning. The form of the survival function for this is
\[S(t)=\exp(-\theta F_0(t))\] for
\(t \geq 0\) where \(F_0(t)\) is a continuous cumulative
distribution function for a non-negative random variable with \(F_0(0)=0\) and \(F_0(t)\uparrow 1\) as \(t\uparrow \infty\). We note that as \(t\uparrow \infty\), \(S(t)\downarrow \exp(-\theta)=c\) where we
will refer to \(c\) as the cure rate.
The function p_pm() assumes \(F_0(t)=1-\exp(-\lambda t)\) is an
exponential cumulative distribution function resulting in the survival
distribution for \(t \geq 0\):
\[S(t; \theta, \lambda) =
\exp(-\theta(1-\exp(-\lambda t))).\] Note that we set the default
of lower.tail=FALSE so that the survival function
computation is the default:
We plot this with \(\lambda = \log(2) / 10\) to make \(F_0(t)\) an exponential distribution with a median of 10. We set \(\theta = -\log(.4)\) to obtain a cure rate of 0.4. we then overlay with the piecewise exponential approximation.
lambda <- log(2) / 10
theta <- -log(.4)
times <- 0:40
plot(times, p_pm(times, theta, lambda), type = "l", ylab = "Survival", xlab = "Time", log = "y")
# Now compute piecewise exponential approximation
x <- seq(8, 40, 8)
pm_rate <- s2pwe(
times = x,
survival = p_pm(x, theta = theta, lambda = lambda)
)
# Now plot the piecewise approximation using the 7-point approximation from above
lines(
c(0, x),
ppwe(x = c(0, x), duration = pm_rate$duration, rate = pm_rate$rate),
col = 2
)
points(c(0, x), p_pm(c(0, x), theta, lambda))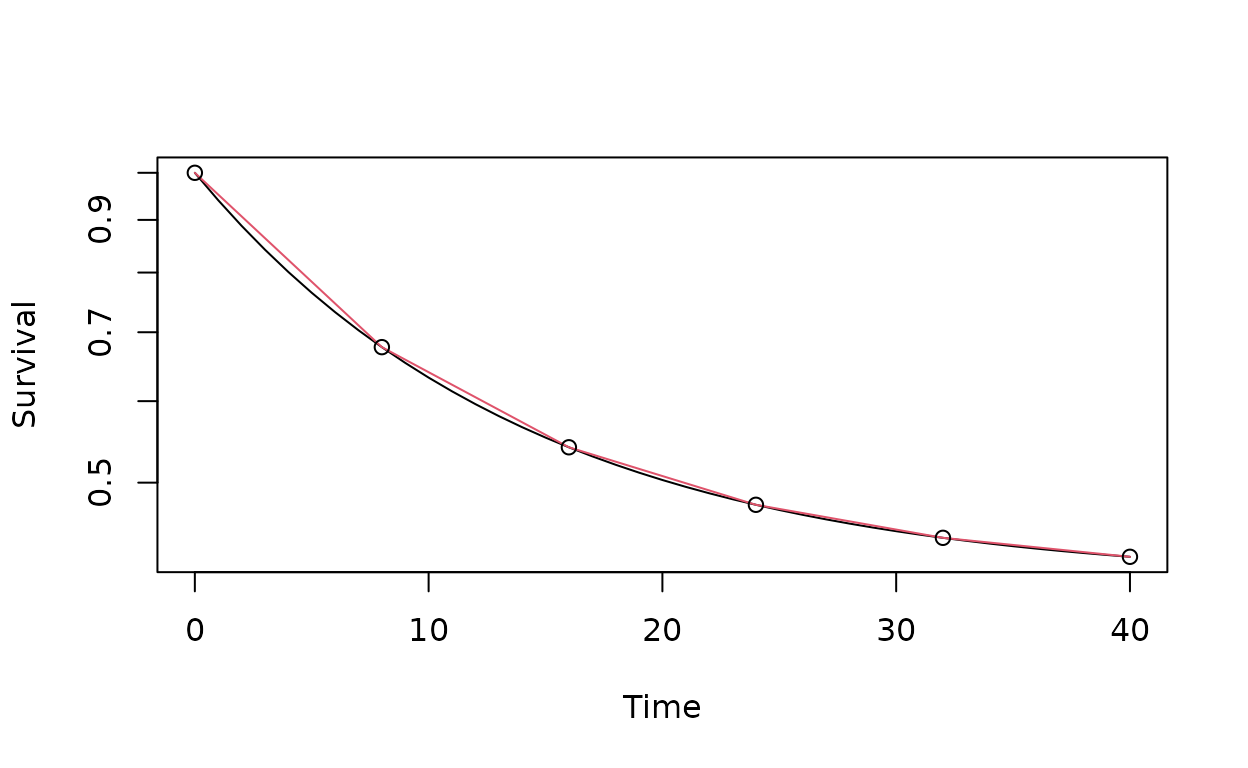
We note that two different \(\theta\) values provide a proportional hazards model in that the ratio of cumulative hazard function \(H(t; \theta, \lambda) = \theta\exp(-\lambda t)\) is constant:
\[\frac{\log(S(t; \theta_1, \lambda))}{\log(S(t; \theta_2, \lambda))} = \theta_1/\theta_2.\]
For a given \(\theta\) value we can compute \(\lambda\) to provide a survival rate \(c_1 > \exp(-\theta)\) at an arbitrary time \(t_1>0\) by setting:
\[\lambda = -\log\left(\frac{\theta - \log(c_1)}{\theta}\right)/t_1.\]
We compute \(\theta\) and \(\lambda\) values with a cure rate of 0.4 and a survival rate of 0.6 at 30 months:
We confirm the survival at time 30:
p_pm(30, theta, lambda)## [1] 0.6