forestly_adsl$TRTA <- factor(
forestly_adsl$TRT01A,
levels = c("Xanomeline Low Dose", "Placebo"),
labels = c("Low Dose", "Placebo")
)
forestly_adae$TRTA <- factor(
forestly_adae$TRTA,
levels = c("Xanomeline Low Dose", "Placebo"),
labels = c("Low Dose", "Placebo")
)
meta <- meta_forestly(
dataset_adsl = forestly_adsl,
dataset_adae = forestly_adae,
population_term = "apat",
observation_term = "wk12",
parameter_term = "any;rel;ser"
) |>
prepare_ae_forestly() |>
format_ae_forestly()
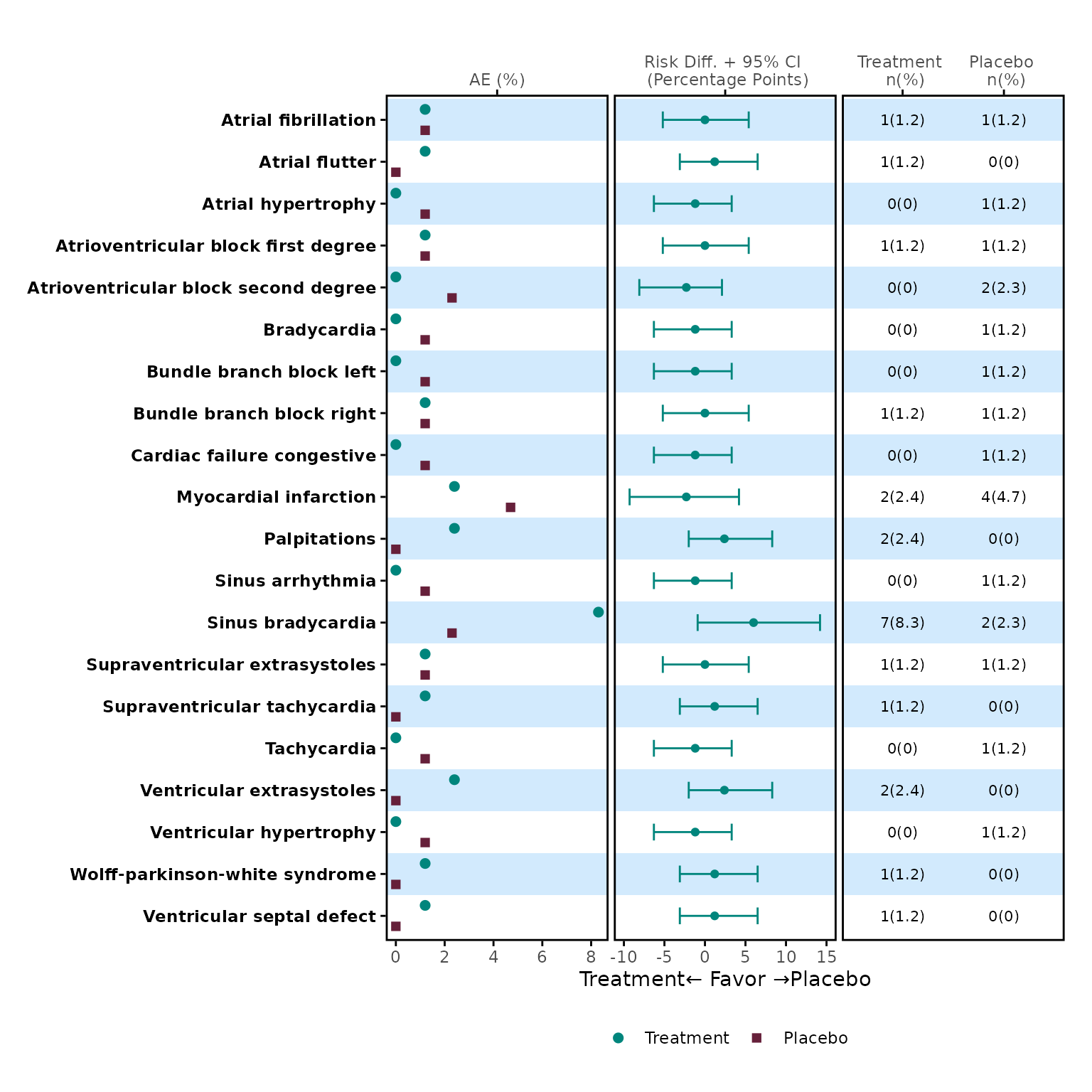
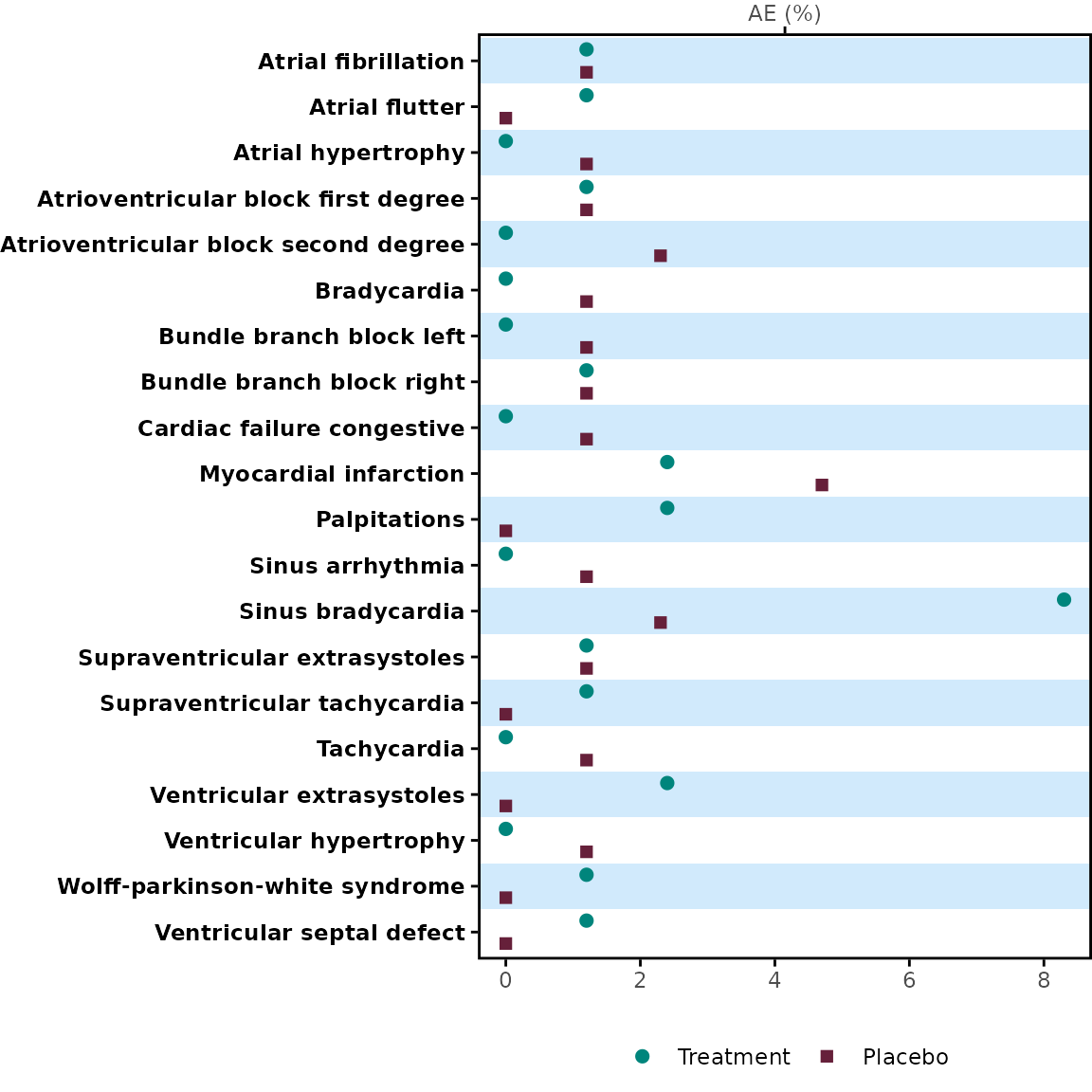
meta_any <- meta$tbl[1:20, ] |> dplyr::filter(parameter == "any")
p1 <- meta_any |>
dplyr::select(name, prop_1, prop_2) |>
plot_dot(
y_var = "name",
label = c("Treatment", "Placebo")
)
#> Warning: The `size` argument of `element_rect()` is deprecated as of ggplot2 3.4.0.
#> ℹ Please use the `linewidth` argument instead.
#> ℹ The deprecated feature was likely used in the forestly package.
#> Please report the issue at <https://github.com/Merck/forestly/issues>.
#> This warning is displayed once per session.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
p1
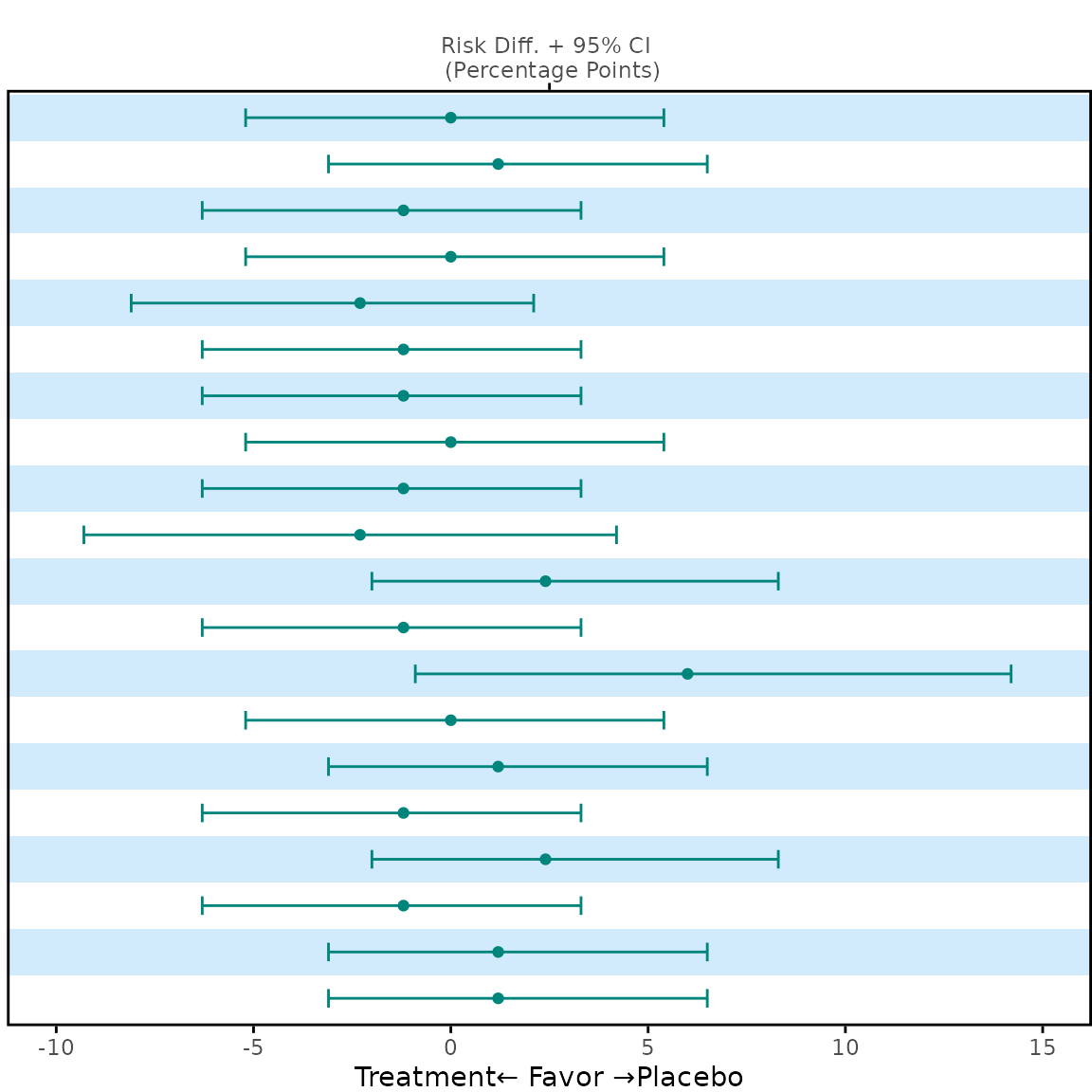
p2 <- meta_any |>
dplyr::select(name, diff_1, lower_1, upper_1) |>
plot_errorbar(
y_var = "name",
label = c("Treatment", "Placebo"),
legend_nrow = NULL,
theme = theme_panel(show_ticks = FALSE, show_text = FALSE)
)
p2
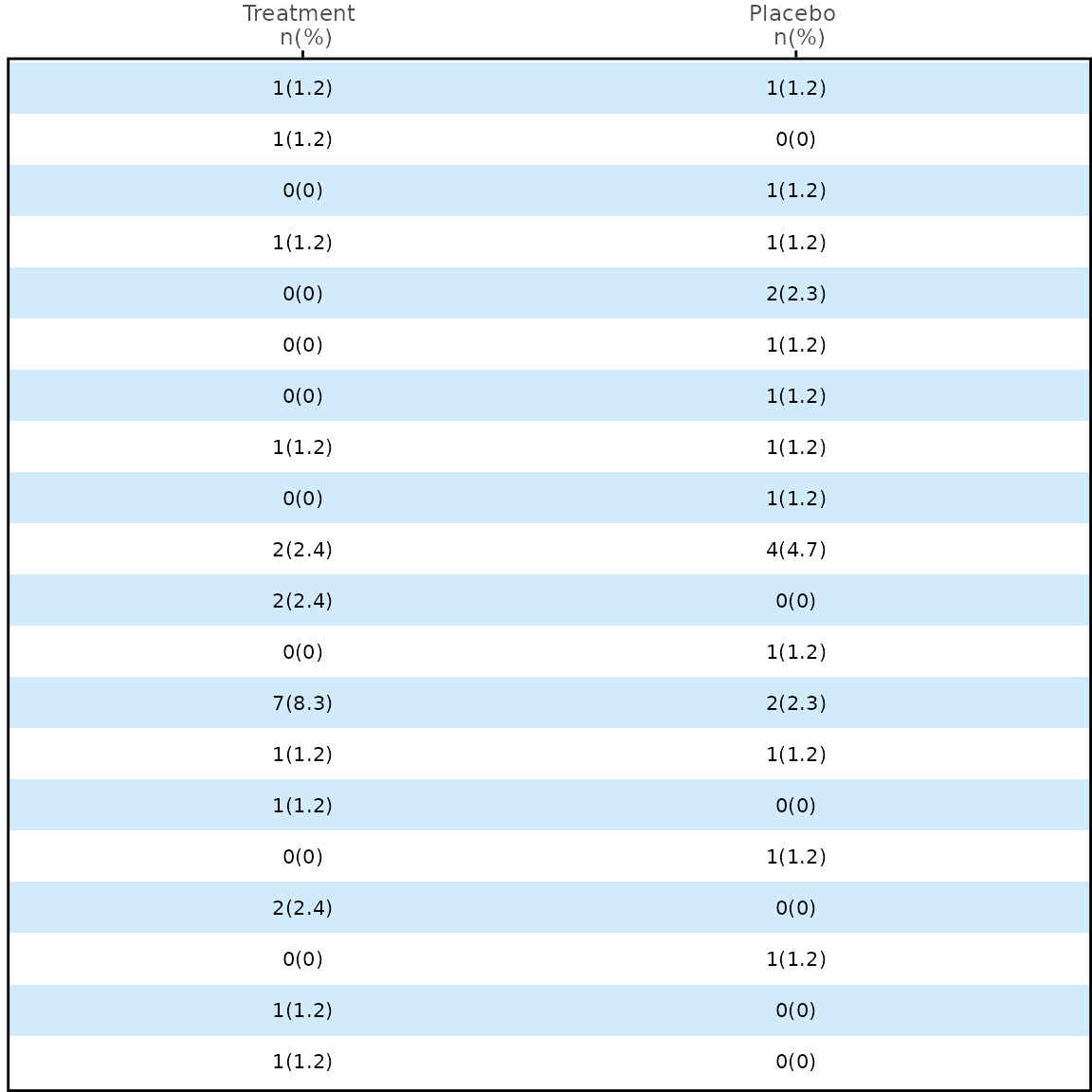
meta_any$stat_1 <- paste0(meta_any$n_1, "(", meta_any$prop_1, ")")
meta_any$stat_2 <- paste0(meta_any$n_2, "(", meta_any$prop_2, ")")
p3 <- meta_any |>
dplyr::select(name, stat_1, stat_2) |>
table_panel(
y_var = "name",
theme = theme_panel(show_ticks = FALSE, show_text = FALSE),
x_label = c("Treatment \n n(%)", "Placebo \n n(%)")
)
p3